Epidiverse

EpiDiverse was a Marie Skłodowska-Curie Innovative Training Network from 1 September 2017 - 28 February 2022. EpiDiverse aimed at the study of epigenetic variation in wild plant species. The network joined research groups from ecology, molecular (epi)genetics and bioinformatics to explore the genomic basis, molecular mechanisms and ecological significance of epigenetic variation in natural plant populations.
The cross-disciplinary research program applied epigenomic research tools to a selection of different wild plants (annual plants, asexually reproducing perennial plants, and long-lived trees) to investigate how epigenetic mechanisms contribute to natural variation, stress responses and long-term adaptation of plants. Understanding the epigenetic contribution to adaptive capacity will help to better understand species responses to global environmental change, and can open new directions for sustainable agriculture and crop breeding.
The EpiDiverse consortium trained 15 Early Stage Researchers to become expert plant epigeneticists, and equipped them with the interdisciplinary skills to successfully tackle this new research area. EpiDiverse training emphasized fluency in both empirical and informatics skills to become creative in working with big ‘omics data in natural contexts.
EpiDiverse is funded by the EU Horizon 2020 programme and involves 12 partners from academia, non-profit organizations and industry located in the Netherlands, Germany, France, Spain, Czech Republic, Italy and Austria.
About The EpiDiverse network
EpiDiverse is a Marie Skłodowska-Curie Innovative Training Network aimed at the study of epigenetic variation in wild plant species. The network joins research groups from ecology, molecular (epi)genetics and bioinformatics to explore the genomic basis, molecular mechanisms and ecological significance of epigenetic variation in natural plant populations.
The cross-disciplinary research program applies epigenomic research tools to a selection of different wild plants: annual plants, asexually reproducing perennial plants, and long-lived trees. Understanding the epigenetic contribution to adaptive capacity will help to better understand species responses to global environmental change, and can open new directions for sustainable agriculture and crop breeding.
EpiDiverse involved academic research groups, non-profit organizations and industry located in the Netherlands, Germany, France, Spain, Czech Republic, Austria, and Italy.
- Netherlands Institute of Ecology (NIOO-KNAW)
Department Terrestrial Ecology
Network Coordinator
https://nioo.knaw.nl/en/department-terrestrial-ecology - Philipps Universität Marburg
Department "Allgemeine Ökologie und Tierökologie"
https://www.uni-marburg.de/fb17/fachgebiete/oekologie/tieroekologie/inde... - Philipps Universität Marburg
Department of Plant Cell Biology
https://www.uni-marburg.de/fb17/fachgebiete/zellbio/zellbioii/home-rensi... - University of Tuebingen
Institute of Evolution and Ecology (EvE)
https://www.uni-tuebingen.de/en/faculties/faculty-of-science/departments... - Botanicky Ustav
Department of population Ecology
http://www.ibot.cas.cz/en/scientific-groups-and-laboratories/department-... - Agroscope
https://www.agroscope.admin.ch/agroscope/en/home.html - DIADE-IRD
http://www.diade-research.fr/en/pages/links/links.html - EcSeq
https://www.ecseq.com/ - EBD-CSIC
www.ebd.csic.es/ - IGA-TS
http://igatechnology.com/ - Martin-Luther-Universität Halle-Wittenberg
Naturwissenschaftliche Fakultät III; Institut für Informatik
http://www.informatik.uni-halle.de/ - Gregor Mendel Institute of Molecular Plant Biology
https://www.gmi.oeaw.ac.at/ - Max Planck Institute for Developmental Biology
https://www.mpg.de/151769/entwicklungsbiologie
EpiDiverse research aims
The capacity of plants to adapt to their environments is key to their success in a world that is constantly changing. Individual plants can adjust to environmental change by being flexible in the traits that they express in different environments (phenotypic plasticity). Over longer time scales populations can adapt to specific environments by selection on heritable traits, resulting for instance in populations with modified flowering time, drought tolerance, or different capacity for phenotypic plasticity. Epigenetic mechanisms play an important role in these processes. Epigenetic mechanisms are reversible chemical modifications of the DNA code that determine if the underlying DNA code is active or silenced. These mechanisms can play a role in the adjustment of gene expression in response to specific environments. Moreover, epigenetic variants exist in plants that are stable between generations, which can lead to differences in heritable traits even between genetically uniform plants.
Thus, to understand the adaptive capacity of plants, we need to look not only at the information within the DNA code, but also at the epigenetic information on top of the DNA code. Much of our current knowledge of DNA methylation in plants comes from research on the molecular model plant species Arabidopsis thaliana. But it is unclear how important epigenetic variation is in the wild for the adaptive capacity of natural plant populations in various ecological contexts. To what extent do epigenetic responses control ecologically relevant stress responses and heritable traits? Do patterns of epigenetic variation in natural populations reflect an important epigenetic role in adaptive capacity? And is the role of epigenetics different for species that differ in important life history traits, such as sexual versus clonal reproduction or in annual versus long-lived species? These are the research questions that EpiDiverse takes on.
The EpiDiverse research applies high-resolution epigenomic research tools and ecological experimental designs to an ecologically diverse set of natural plant species, with the ultimate aim to expose the contribution of epigenetic variation to the adaptive capacity of plants.
Specific research objectives of the network are to:
- Optimize genomic and bioinformatic tools to enable high-resolution DNA methylation analysis in non-model plant species
- Identify natural patterns of DNA methylaiton variation and its associations with phenotype, local environment, climate and geographic location
- Unravel the underlying molecular mechanisms of epigenetic responses to ecologically relevant stress environments.
Project structure
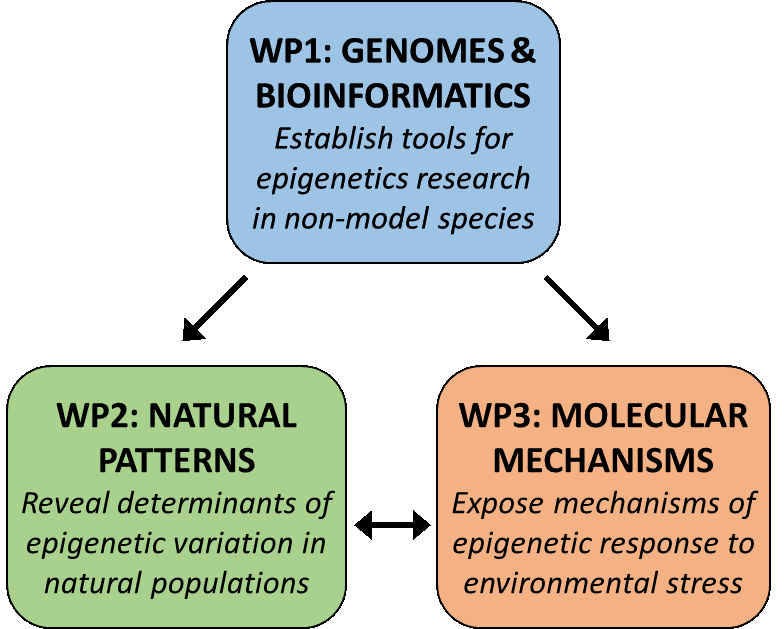
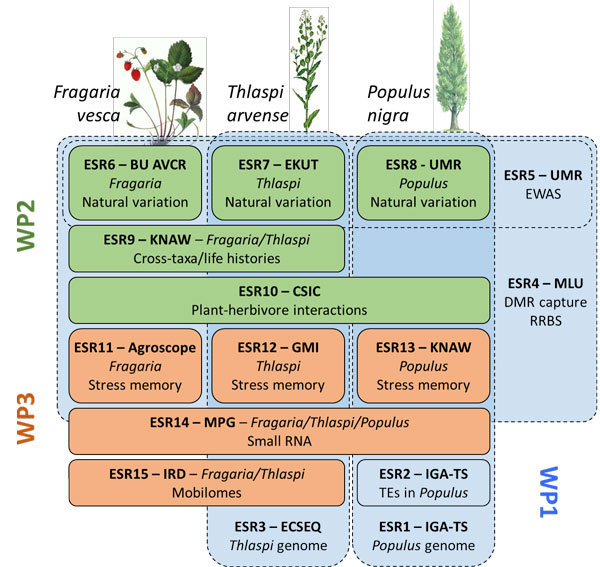
In EpiDiverse, research projects are clustered around a few study species (Populus nigra, Fragaria vesca and Thlaspi arvense) with very different life histories and ecological characteristics. For each species, research groups from different backgrounds collaborate to generate genomic resources and epigenomic analysis pipelines (the “bioinformaticians”, work package 1) and to use these tools for exploring patterns of epigenetic variation in natural populations (the “ecologists”, work package 2) and to understand the epigenetic mechanisms of plant responses to environmental stress (the “molecular biologists”, work package 3).
Projects
We have 15 PhD projects in 3 work packages. PhD students from the fields of plant sciences, ecology, (epi)genetics, (epi)genomics, computational biology or bioinformatics work on these projects. The EpiDiverse network facilitates close collaboration between individual PhD projects. These projects provide an excellent opportunity to receive cross-disciplinary doctoral training and to do cutting-edge research in plant epigenetics.
The EpiDiverse research focuses on three wild plant species. For each of these species, research groups from computational biology, molecular genetics and ecology team up to address the genomic basis, the molecular mechanisms and the ecological significance of epigenetic variation.
EpiDiverse provides a comprehensive training program that includes summer schools, workshops and exchange visits to different groups in the network. We will offer training in molecular, genomic, computational, and ecological aspects of studying epigenetic variation, as well as in transferable skills. Students will be part of a group of 15 PhD students from across Europe that will all work on similar questions, and that will regularly meet and interact.
Projects
-
- PhD Supervisor(s): Michele Morgante, Emanuele De Paoli, Federica Cattonaro
- Host Institution: Institute of Applied Genomics (IGA) - Technology Services, Udine, Italy
- Duration: 36 months.
- Fixed start date: 1 April 2018
- Planned secondment(s): ecSeq GmbH (DE), Philipps Universitaet Marburg (DE)
The aim of this project is to produce a high quality, annotated reference sequence of the Populus nigra (black poplar) genome in order to support structural and functional studies as well as enable high-resolution epigenetic analyses within the EpiDiverse consortium. Genome assembly will be informed by the available draft genome of a sister species, Populus trichocarpa and aimed at producing haplotype-specific assemblies for each of the two homologs present in the highly heterozygous individual chosen for sequencing. DNA/RNA library preparation and sequencing will be performed under the supervision of highly specialized staff in the laboratories of the hosting sequencing facility using long pseudo-read Next Generation Sequencing technologies. The application of state-of-the-art in silico analyses and experimentation of new computational methods, some of which developed in house, will yield an initial whole-genome draft that will be further assembled into large scaffolds using in situ Hi-C technologies. The phased assemblies of the two parental haplotypes will allow analysis of the intra-individual structural variation and will be employed to enable allele-specific analyses of gene expression and epigenetic determinants. The genome sequence will be further refined and completed with the addition of gene prediction and repeat element annotation. A specific focus will be placed on the analysis of the transposable element composition and the study of their evolution and insertional preferences using also existing sequencing data from additional Populus nigra accessions. The project will interact with other international research teams involved into the exploitation of poplar genomic information to explore ecologically relevant phenomena.
More information
-
- PhD Supervisor(s): Michele Morgante, Emanuele De Paoli, Federica Cattonaro
- Host Institution: Institute of Applied Genomics (IGA) -Technology Services, Udine, Italy
- Duration: 36 months.
- Fixed start date: 1 April 2018
- Planned secondment(s): Philipps Universitaet Marburg (DE), Institut National de la Recherche Agronomique (FR)
Different combinations of epigenetic marks mediate the timely expression of genetic determinants and the silencing of detrimental repetitive elements. However, the epigenetic suppression of transposable elements (TEs) can accidentally impact the transcriptional status of neighbouring genes generating epigenetic variants (obligatory or facilitated epialleles) associated with differential phenotypes. To date, these cases provide the most convincing examples of naturally occurring epialleles genetically transmitted by plants. Thus, the capability of environmental stresses to alter the epigenomic landscape and trigger TE activity raises a compelling theme of research focusing on the potential role of TEs in mediating the regulation of gene expression by environmental cues.
In this project we aim to explore the three-way relationship potentially existing between the DNA methylome of poplar (Populus nigra), the highly dynamic transposable element systems that represent a major contributor of plant genomic structural variation and the environmental challenges plants are exposed to. Through the integration of Next Generation Sequencing approaches, single nucleotide polymorphism analysis and genomic maps of structural variants, we aim to produce allele-specific single-base resolution maps of the DNA methylome in poplar emphasizing the effects of the most variable and repetitive component of the genome. The resulting data will be combined with DNA methylation screening in common garden experiments to identify the most reactive TE families and unveil the epigenetic variation that may link TE silencing to gene regulation in an ecologically relevant manner.
More information
-
- PhD Supervisor(s): David Langenberger, Peter Stadler
- Host Institution: ecSeq Bioinformatics GmbH, Leipzig, Germany
- Duration: 36 months.
- Fixed start date: 1 April 2018
- Planned secondment(s): Gregor Mendel Institute of Molecular Plant Biology (Vienna, Austria), IGA Technology Services SRL (Udine, Italy) and Roche Diagnostics GmbH (Mannheim, Germany)
DNA methylation variants can arise spontaneously, they can be under genetic control or they can be induced by the environment. In plants, some DNA methylation variants are stable across many generations whereas other variants are very transient. A good understanding of the transgenerational dynamics of DNA methylation variants is essential to understand their impact on heritable traits and their effect on adaptation. Current insight in the transgenerational dynamics of DNA methylation is limited to very few model plant species, but it is predicted that these dynamics are not constant among different plants. For instance, plant reproduction mode can have a large effect because asexual reproduction bypasses some of the epigenetic resetting mechanisms that normally occur during sexual reproduction. Adaptive differences in transgenerational stability may also differ between species with different life spans or from habitats of different environmental predictability.
In this project, we aim to investigate differences in DNA methylation dynamics between species with different life history traits. In close collaboration with other bioinformaticians and biologists, a best-practice pipeline for the analysis of plant bisulfite data will be developed, benchmarked, and provided to be applied to the Next-Generation Sequencing (NGS) data generated in the frame of different EpiDiverse projects. In addition, novel algorithms will be devised and implemented to better detect DNA methylation variants and search for important methylation haplotypes to better understand the epigenetic regulation. The special constellation of working at a bioinformatics company in very strong cooperation with the chair of bioinformatics at the university offers the unique possibility of gaining benefits from both: The clearly structured style of the work in a company will help implementing novel ideas from academia in the light of recent publications, resulting in a strong focus and high productivity.
More information
-
- PhD Supervisor(s): Ivo Grosse, Carolin Delker, Peter Stadler
- Host Institution: Martin Luther University Halle-Wittenberg
- Duration: 36 months.
- Fixed start date: 1 April 2018
- Planned secondment(s): EcSeq Bioinformatics Leipzig (GE), NIOO-KNAW Wageningen (NL), Roche Diagnostics Germany
Bioinformatics plays a central and highly integrative role in the EpiDiverse network. The scientific goal of this project is the development and optimization of algorithms and pipelines (i) for the computational analysis of whole-genome bisulfite sequencing (WGBS) data, reduced-representation bisulfite sequencing (RRBS) data, as well as high-throughput methylation data obtained from novel technologies developed by the EpiDiverse network, (ii) for the computational recognition of sequence patterns using modern machine learning techniques, (iii) for detecting, quantifying, and visualizing correlations among these patterns and of these patterns with functional annotations, and (iv) for the detection, quantification, and visualization of genetic as well as epigenetic population structure in diverse datasets generated by the EpiDiverse network.
More information
-
- PhD Supervisor(s): Stefan Rensing & Noe Fernandez-Pozo
- Host Institution: Philipps-University Marburg, Marburg, Germany
- Duration: 36 months.
- Fixed start date: 1 April 2018
- Planned secondment(s): EcSeq Bioinformatics (https://www.ecseq.com/) for joint bioinformatics pipeline development, Max Planck Gesellschaft (MPI Tübingen) for training in DMR and GWAS/EWAS methods, and Roche Diagnostics Germany for learning about new DNA methylation screening methods.
The successful candidate will be trained on state-of-the-art bioinformatics and epigenetics methods. He or she will participate in an exciting and innovative project to develop a set of pipelines and tools for epigenomics data analysis on non-model plants. The main task will be the development of a pipeline facilitating epigenome-wide association studies for EpiDiverse. The developed software will be applied to study a large set of epigenetic data produced to understand the epigenetic changes happening in plants from different natural populations, how these changes are inherited after sexual and asexual reproduction and how they differ in annual and perennial species such as Fragaria vesca (strawberry) and Populus nigra (black poplar).
More information
-
- PhD Supervisor(s): Vítek Latzel, Zuzana Münzbergová
- Host Institution: Institute of Botany of the Czech Academy of Sciences, Průhonice near Praha, Czech Republic
- Duration: 36 months with possible extension for another 12 months.
- Fixed start date: 1 April 2018
- Planned secondment(s): ecSeq Bioinformatics Leipzig (GE); Philipps Universitaet Marburg (GE); Institute National de la Recherche Agronomique, Angers (FR)
Plant trait variation is commonly assumed to be driven by DNA sequence variation. However, increasing body of evidence suggests that epigenetic variation such as DNA methylation can be additional mechanism that enables heritable trait variation. Epigenetic variation can be induced by environment and can be independent of genetic variation. Epigenetic variation may thus be an important factor in adaptation of natural populations that can operate at faster time scale than adaptation based on DNA sequence variation.
In this project, we will evaluate patterns of epigenetic variation of natural populations of wild strawberry (Fragaria vesca) along climatological gradient. We will screen methylation variants of samples taken from plants of natural populations and samples of the same clones that have been transferred to a common garden environment. This will allow characterizing epigenetic population structure, association of methylation variants with relevant ecological and climatic variables and comparison of epigenetic population structure determined by environmental induction relative to stably inherited methylation variation. In addition, we will evaluate the role of DNA methylation variation in local adaptation by conducting reciprocal transplant experiments using experimentally demethylated and naturally methylated plants. In cooperation with members of the EpiDiverse network we will also focus on the role of transposable elements in population differentiation and their responses to environmental stress.
More information
-
- PhD Supervisor(s): Oliver Bossdorf, Niek Scheepens
- Host Institution: Eberhard Karls University of Tübingen, Tübingen, Germany
- Duration: 36 months
- Fixed start date: 1 April 2018
- Planned secondment(s): ecSeq Bioinformatics, Leipzig (GE); Gregor Mendel Institute, Vienna (AT); Philips University Marburg (GE)
There is currently much speculation about the extent, stability and ecological relevance of epigenetic variation in natural plant populations. How much epigenetic variation is there, and how is it structured? Do patterns of epigenetic variation mirror those of genetic variation, or do they tell a different story? How much of the variation observed in the field is plastic responses to different environments, and how much is stably inherited and thus potentially adaptive? So far, we cannot answer these questions well enough because there have been too few systematic large-scale surveys of epigenetic variation in natural plant populations.
The aim of this research project will be to conduct such a study in the annual plant and common crop weed Thlaspi arvense (field pennycress). The PhD student will take part in a sampling campaign across Europe and set up a garden experiment, will screen DNA methylation in field and garden plants to assess epigenetic population structure and the stability of DNA methylation differences, and will analyse relationships between epigenetic variation, climate and other environmental factors. The project will be carried out in close collaboration with the group of Claude Becker at the Gregor Mendel Institute in Vienna (RP12).
A unique aspect of this project is the breadth of its methods and of the training that the student will receive. Besides the training in field and experimental ecology, and the training opportunities through the central EpiDiverse summer schools, there will be several intensive training stays (secondments) with different EpiDiverse partners to learn modern bioinformatic and statistical methods for epigenome analyses.
More information
-
- PhD Supervisor(s): Lars Opgenoorth & Katrin Heer
- Host Institution: Philipps-Universität Marburg, Marburg, Germany
- Duration: 36 months
- Fixed start date: 1 April 2018
- Planned secondment(s): EcSeq Bioinformatics Leipzig (GE) (https://www.ecseq.com/) for WGBS data analysis, NIOO-KNAW Wageningen (NL) for learning methylation screening library preparation.
Trees as long-lived and sessile organisms are particularly challenged by climate change. Thus epigenetic changes via priming either during embryogenesis or at a later life stage might be particularly relevant for long-lived plant species, independent of whether these changes are heritable (generative/adaptation) or act during the lifetime (somatic/acclimation) only. Thus, studying epigenetic processes in trees might crucially contribute to our understanding of the strategy of these long-lived organisms for coping with environmental change and might help explain their ecological and evolutionary success. In this project, DNA methylation and TE variants will be screened in natural Populus nigra populations and Populus nigra italica clones that have been growing in natural environments along European-scale geographic and climatological gradients. Specifically we will characterize epigenetic population structure, associate methylation and TE variants with relevant ecological and climatic variables, and assess the contribution to epigenetic population structure of environmental induction relative to stably inherited methylation variation. DNA methylation and TE screening tools will include whole genome bisulfite sequencing, and targeted bisulfite sequencing. The project will interact closely with bioinformatics groups as well as stress experimental groups in the EpiDiverse network.
Contact information
-
- Supervisor(s): Koen Verhoeven & Wim van der Putten
- Host Institution: Netherlands Institute of Ecology (NIOO-KNAW), Wageningen, The Netherlands
- Duration: 48 months
- Fixed start date: 1 April 2018
- Planned secondment(s): Martin Luther University Halle-Wittenberg, Halle (GE); Institute National de la Recherche Agronomique, Angers (FR); Wissenschaft im Dialog, Berlin (GE)
DNA methylation variants can arise spontaneously, they can be under genetic control or they can be induced by environments. In plants, some DNA methylation variants are stable across many generations whereas other variants are very transient. A good understanding of the transgenerational dynamics of DNA methylation variants is essential to understand their impact on heritable traits and their effect on adaptation. Current insight in the transgenerational dynamics of DNA methylation is limited to very few plant species, but it is predicted that these dynamics are not constant between plant species. For instance, plant reproduction mode can have a large effect because asexual reproduction bypasses some of the epigenetic resetting mechanisms that normally occur during sexual reproduction. Adaptive differences in transgenerational stability may also differ between species with different life spans or from habitats of different environmental predictability.
In this project, we aim to investigate differences in DNA methylation dynamics between species with different life history traits. Specifically, we will characterize environmental effects and transgenerational stability of DNA methylation and compare these between sexually and asexually reproducing wild Fragaria (strawberry) species, and between different Brassicaceae species that differ in life history characteristics (for instance annual versus perennial) and habitat characteristics. DNA methylation screening tools will include reduced representation bisulfite sequencing and the project will interact closely with bioinformatics groups in the EpiDiverse network.
More information
-
- PhD Supervisor(s): Conchita Alonso & Mónica Medrano
- Host Institution: Consejo Superior de Investigaciones Científicas - Estación Biológica de Doñana (EBD-CSIC), Sevilla, Spain
- Duration: 36 months
- Fixed start date: 1 April 2018
- Planned secondment(s): Netherlands Institute of Ecology (NIOO-KNAW), Wageningen, NL; Martin Luther University Halle-Wittenberg, Halle (GE); Wissenschaft im Dialog, Berlin (GE).
Plant species can defend against herbivore damage in different ways. Mounting evidence indicates that epigenetic regulation can modulate such responses, including priming and maternal effects on resistance. Still, our understanding of epigenetic contribution to plant-herbivore interactions is fragmentary.
In this project we aim to evaluate the role of DNA methylation in plant responses to herbivory by screening the effects of herbivory in DNA methylation, resistance traits, and individual performance in the focal species of EpiDiverse. Further, the potential for transgeneration inherited effects and causal involvement of DNA methylation will be experimentally tested under controlled conditions in short-lived species.
More information
-
- Supervisor(s): Etienne Bucher, Béatrice Denoyes
- Host Institution: Agroscope, Changins, Switzerland
- Duration: 36 months
- Fixed start date: 1 April 2018
- Planned secondment(s): Leipzig University (GE), Charles University Prague (CZ), Udine University (IT), Institut de recherche pour le développement (FR)
Transposable elements (TEs) have long been known to contribute to genetic diversity in plants that undergo severe stresses. However, it is currently unknown how and to which level the activity of TEs can contribute to stable epigenetic changes in plants. In this project, we will use several innovative approaches to study heritable genetic and epigenetic changes resulting from the mobilization of transposable elements in strawberry (Fragaria vesca). These approaches will include: (i) identification of ecologically relevant stresses and combinations thereof that lead to the mobilisation of TEs (ii) create strawberry lines carrying novel TE insertions (iii) study the phenotypic diversity and modified stress response of the resulting plants (iv) study how the mobilisation and insertion of novel TEs affect the strawberry epigenome (via whole genome DNA methylation analysis), (v) analyse the stability of stress-induced DNA methylation changes during sexual and asexual reproduction.
To allow detailed molecular and genome-wide characterization of these experiments a simplified genetic strawberry material (near recombinant inbred lines) will be used.
The research groups of E. Bucher and B. Denoyes have strong track record in the study of epigenetics and strawberries (e.g. Daccord et al (2017), Nature Genetics and Tenreira et al (2017), The Plant Cell).
More information
-
- PhD Supervisor(s): Claude Becker, Ortrun Mittelsten Scheid
- Host Institution: Gregor Mendel Institute of Molecular Plant Biology, Vienna, Austria
- Duration: 36 months
- Fixed start date: 1 April 2018
- Planned secondment(s): Institute of Applied Genomics - Technology Services, Udine (IT); Eberhard Karls University Tübingen (GE); University of Leipzig (GE)
Pennycress (Thlaspi arvense) is a member of the Brassicaceae family; it is a selfing diploid that is widespread in Europe, and can be found in natural, agricultural, and disturbed sites. T. arvense is of dual interest to us: on the one hand, as a weed it infests agricultural areas and causes losses in agricultural production. On the other hand, it has recently been established as a promising biofuel crop. Its close relatedness to the well-studied model plant A. thaliana and other well-characterised Brassica species makes it amenable to genomic enquiries at the whole-genome level.
In this project we ask how natural epigenetic variation in T. arvense influences the plant’s response to short- and long-term exposure to abiotic and biotic stress. Objectives are: (1) identifying genomic loci that change their epigenetic configuration upon drought treatment; (2) determining the transgenerational stability of stress-induced changes in dependence of the duration, intensity and frequency of the stress; (3) assessing the contribution of epigenetic stress-regulated loci to the physiological stress response of the plant. The project builds on our year-long expertise in the analysis of whole-genome DNA methylation profiles and is carried out in close collaboration with Oliver Bossdorf at the University of Tübingen, Germany. The PhD student will gain experience in complex experimental design, state of the art whole-genome profiling techniques using next-generation sequencing technology, and computational analysis of whole-genome data.
More information
-
- Supervisor(s): Koen Verhoeven & Wim van der Putten
- Host Institution: Netherlands Institute of Ecology (NIOO-KNAW), Wageningen, the Netherlands
- Duration: 48 months
- Fixed start date: 1 April 2018
- Planned secondment(s): Leipzig University (GE); Philipps-Universität Marburg (GE); Institute of Applied Genomics - Technology Services, Udine (IT)
In long-lived sessile organisms such as trees, phenotypic plasticity is an important requirement for successful persistence in changing or variable environments. Epigenetic mechanisms have the potential to mediate long-term plastic responses to environmental change. However, the importance of epigenetic mechanisms such as DNA methylation as regulators of adaptive plasticity is not well known. In this project we will experimentally evaluate effects of stress exposure on DNA methylation, transposable element activity and gene expression in black poplar (Populus nigra), with the aim to (1) identify genomic loci that show stress-induced epigenetic modification with functional consequences, and (2) evaluate the temporal stability of such loci, for example across growing seasons. Making use of clonally propagated trees that have grown in contrasting environments, we will also investigate to what extent environment-induced epigenetic differences are transmitted to offspring via clonal propagation (cuttings) versus via sexual reproduction through seeds, using allele-specific DNA methylation analysis. Experiments will be carried out in close interaction with other Populus nigra projects within the EpiDiverse consortium (at Marburg University, Germany and at the Institute of Applied Genomics, Udine, Italy).
More information
-
- PhD Supervisor(s): Detlef Weigel & Anjar Wibowo
- Host Institution: Max Planck Institute for Developmental Biology, Tübingen, Germany
- Duration: 36 months
- Fixed start date: 1 April 2018
- Planned secondment(s): Institut de Recherche pour le Developpement (FRA), IGA Technology Services (ITA)
Epigenetic mechanisms can establish a memory of previous stress exposure, allowing a plant to react more effectively upon repeated exposure to the same stress. In some cases, such memory may extend to the following generation.
Prominent targets of stress-induced epigenetic changes are transposable elements. Their induced methylation can repress neighboring genes, and thus modify cellular output and stress response. Such changes may involve stress-induced small RNAs generated from and/or recognizing sequences found in transposable elements.
Objective is to study the transgenerational aspect of epigenetic memory by integrating information on small RNA abundance and DNA methylation both within and between plant generations. Of particular interest will also be DNA sequence variation among related genotypes of the same species and its effects on stress memory. This may be caused by the absence of transposable elements, the absence of transposon repression through altered small RNA sequences, misregulation of small RNA abundances, or additional components of epigenetic control. Next generation sequencing, including data analysis, will be an integral part of the project.
More information
- Website Weigelworld and Max Planck Institute
- Project video
-
- PhD Supervisor(s): Marie Mirouze & Alain Ghesquière
- Host Institution: Institut de recherche pour le développement (IRD Diade), Monpellier, France
- Duration: 36 months
- Fixed start date: 1 April 2018
- Planned secondment(s): IGA-TS (Udine, Italy) to receive training in TE-epiallele, MPG (Max Planck, Tübingen, Germany) for integration of sRNA with TE activity data.
Transposable elements (TEs) represent a main source of genomic diversity and an evolutionary force in plants. Host genomes have developed epigenetic mechanisms to control and prevent their proliferation. However, under specific stress conditions or at precise developmental stages, some TEs can be remobilized and proliferate in plant genomes. As TEs attract efficiently repressive epigenetic marks their mobility can have an impact on nearby gene regulation. In plants, only a few active TEs have been identified and the mobile part of the genome or mobilome, comprising these elements, is unknown. Notably the extent of TE mobility in plant populations in the wild is not known. To establish an unbiased repertoire of mobile TEs during plant development and in ecological conditions, we have developed the mobilome-seq as a simple strategy based on high throughput sequencing to detect TEs extrachromosomal forms (ecDNAs). It represents a novel approach to understand and evaluate the extent and impact of real time TE mobility on eukaryotic genomes.
In this project, the objectives are (1) to identify the full repertoire of mobile TEs through mobilome sequencing in stressed plants as compared to control plants, (2) to analyze the impact of identified TE mobility on genome stability and on epigenetic gene regulation. A pooled-individual mobilome-seq strategy will be used to identify treatment-specific ecDNAs. Subsequently, candidate mobile TEs detected by mobilome-seq will be screened using a sequence capture approach to evaluate neo-insertion sites. The impact of neo-insertions on gene regulation and epigenetic control will be assessed. We will compare the mobilomes of sexually and asexually reproducing wild Fragaria (strawberry) species, and of different Brassicaceae species that differ in life history characteristics (annual versus perennial) and habitat characteristics. On the same material, methylome and small RNA sequencing data will be made available by other groups from the EpiDiverse network and will be implemented in the analysis of TE candidates.
The PhD student will gain experience in computational analysis of genomic data and molecular biology (DNA extraction, ecDNA amplification, sequence capture).
More information
EpiDiverse training aims
Interdisciplinary scientific training
The research program of EpiDiverse was characterized by integration of bioinformatics, molecular genetics and ecology expertise. A key aim of the academic training provided to EpiDiverse students was to develop the interdisciplinary skills for combining these fields. The ability to link genetic/epigenetic information with experimental proficiency, bioinformatics skills, and creativity in distilling knowledge from big data. This training prepared for academic careers in modern biology where technological ‘omics’ advancements are revolutionizing genetic, ecological, and medical research agendas.
Experience in non-academic work environments
The EpiDiverse consortium includes academic and non/academic participants. The non-academic participants carried out an integral part of the research program and host several of the EpiDiverse research projects. Furthermore, they provided training opportunities (secondments) for those students that were hosted at academic research groups in the network. Non-academic participants were:
- Roche Diagnostics, Mannheim, Deutschland GmbH. At Roche the students can experience the industrial work environment. Here the bioinformatics students will be trained in new technology for DNA methylation screening.
- EcSeq contributes with knowledge of pipeline maintenance, hands-on bioinformatics training. This company supported the students with their data analysis.
- IGA-TS, IGA Technology Services SRL, contributed with expertise of (Epi)genomic sequencing data and facilities for library preparation and Illumina sequencing.
- All three mentioned companies, ECSEQ, IGA-TS and ROCHE, represent large and growing life sciences and -omics data industries.
- WiD (Wissenschaft im Dialog), Berlin, Germany trained the students in public outreach of information and science communication.
Transferable skills
EpiDiverse organized platforms for scientific presentations and interaction, training in research ethics, data management, scientific writing and project design to ensure that all ESRs received training in these essential research skills. Because the research topic of EpiDiverse is of fundamental scientific interest, it was important for the ESRs to develop skills in science communication in order to explain the research and its relevance to a general audience. ESR received training in science communication and helped develop an online portal for learning on epigenetics, which was integrated with the outreach goals of the network
Previous trainings
-
The school aimed to establish a common knowledge base in ecological epigenetics concepts for students from all three disciplines and to inspire students to tackle big questions in plant ecological epigenetics and epigenomics. Training in this school provided the foundation for the ESRs to internalize the multi-disciplinary EpiDiverse research agenda. In addition, workshops were given to help the students to successfully manage their PhD projects.
-
This Summer School was an extended hands-on bioinformatics bootcamp. The school aimed to provide an understanding and hands-on experience in Next-Generation Sequencing data analysis with a special focus on bisulfite sequencing and is co-organized by the bioinformatics groups within EpiDiverse that developed and benchmarked DNA methylation analysis pipelines. This school made analysis approaches available to all ESRs and train the ESRs on the concepts and the practical use of the centralized EpiDiverse high-performance cluster; and enable ESRs to independently use best-practices bisulfite sequencing analysis pipelines and data archiving protocols.
-
During this Summer School the ESRs got specialized training in methods for downstream analyses that combined (epi)genetic, environmental, and geographic information to draw ecological insights. These skills were essential for understanding the (epi)genomic responses to ecological variation but they also provide a sound basis for creative use, integration and interpretation of large data in general, which strengthens career opportunities beyond academia in the open data and internet-based economy.
-
This school emphasized transferable skills: science communication and entrepreneurship. There was a science communication workshop. Further the ESRs worked jointly on contributing information from their projects to the EpiDiverse online learning portal. A second focus was on entrepreneurship where partners and beneficiaries will provide insights into careers in life science companies and experiences with founding of a start-up company will be shared with the ESRs. The ESRs were introduced to commercial exploitation of genetic resources in Fragaria and Populus by representatives of commercial companies.
-
Linking Ecology, Molecular Biology and Bioinformatics in Plant Epigenetic Research Sevilla, Spain, 29 september – 1 october 2021
Organized by the European Training Network EpiDiverse (H2020-MSC-ITN-764965)
To mark the end of the EpiDiverse programme an open scientific conference on plant ecological epigenetics and epigenomics was organized Seville in 2021. At this conference, ESRs presented their results with the broader scientific community and transfer knowledge on epigenetic diversity to stakeholders from biodiversity management and conservation.
There is an ever-growing interest in the mechanisms underlying epigenetic phenomena, which may be particularly crucial for the survival of plants within dynamic environments as they cannot respond behaviourally or migrate immediately. Understanding the epigenetic contribution to plant phenotypic variation, stress responses and long-term adaptation will help to better understanding species responses to global environmental change, and can open new directions for sustainable agriculture and crop breeding. Moving forward this field requires multidisciplinary cross-talk between ecologists, molecular geneticists and bioinformaticians. This was the aim of the EpiDiv2021 Conference celebrated at CaixaForum Center in Sevilla (Spain) 29th september - 1st october 2021.
Organizing committee
Conchita Alonso1, Troyee Anupoma1, Francisco Balao2, Claude Becker3, M Teresa Boquete1, Oliver Bossdorf4, Noé Fernández-Pozo5, Katrin Heer5, Mónica Medrano1, Xavier Picó1, Lars Opgenoorth5, Koen Verhoeven6.
1Estación Biológica de Doñana, Consejo Superior de Investigaciones Científicas (CSIC), Spain
2Plant Biology and Ecology Department, University of Seville, Spain
3Gregor Mendel Institute of Molecular Plant Biology, Austrian Academy of Sciences, Vienna BioCenter (VBC), Austria
4Plant Evolutionary Ecology, University of Tübingen, Germany
5Philipps University of Marburg, Germany
6Netherlands Institute of Ecology (NIOO-KNAW), NetherlandsKeynote Speaker
- Carlos M. Herrera, Estación Biológica de Doñana, Consejo Superior de Investigaciones Científicas (CSIC), Spain
Invited Speakers
- Hanne de Kort, Biology Department, KU Leuven, Belgium
- Xiaoqi Feng, John Innes Centre, UK
- Helene Kretzmer, Max Planck Institute for Molecular Genetics, Germany
- Alexander Jueterbock, Faculty of Biosciences and Aquaculture, Nord University, Norway
- Stéphane Maury, University of Orleans, France
- Jurriaan Ton, Department of Animal and Plant Science, University of Sheffield, UK
Contact
- info@epidiverse.eu
- T +31 (0)317 473624
- X EpiDiverse
EpiDiverse Coordinator
Dr. Koen Verhoeven
Department of Terrestrial Ecology
Netherlands Institute of Ecology (NIOO-KNAW)
EpiDiverse Project manager
Margreet Bruins
Department Terrestrial Ecology
Netherlands Institute of Ecology (NIOO-KNAW)
Visiting address
Droevendaalsesteeg 10
6708 PB Wageningen
T 0317 47 34 00
nioo.knaw.nl
Postal address
Postbus 50
6700 AB Wageningen
For members
For consortium member access to archived project documents, please contact Koen Verhoeven